Getting Started#
Installation#
The Read Structure step is probably already installed in your SEAMM environment, but if not or if you wish to check, follow the directions for the SEAMM Installer. The graphical installer is the easiest to use. In the SEAMM conda environment, simply type:
seamm-installer
or use the shortcut if you installed one. Switch to the second tab, Components, and check for read-structure-step. If it is not installed, or can be updated, check the box next to it and click Install selected or Update selected as appropriate.
The non-graphical installer is also straightforward:
seamm-installer install --update read-structure-step
will ensure both that it is installed and up-to-date.
Example#
This simple flowchart download
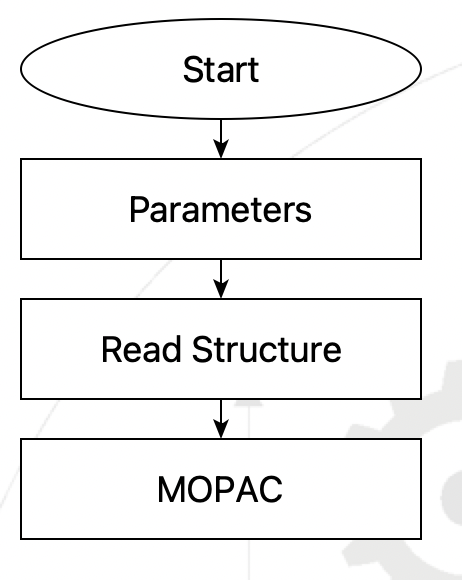
Simple flowchart using Read Structure step#
will read a molecular structure file whose name is given in the Control Parameters step, and optimize the structure using MOPAC.
Editing the step brings up a dialog like this, which takes the filename, the type of file, etc. plus where to put the structure(s):
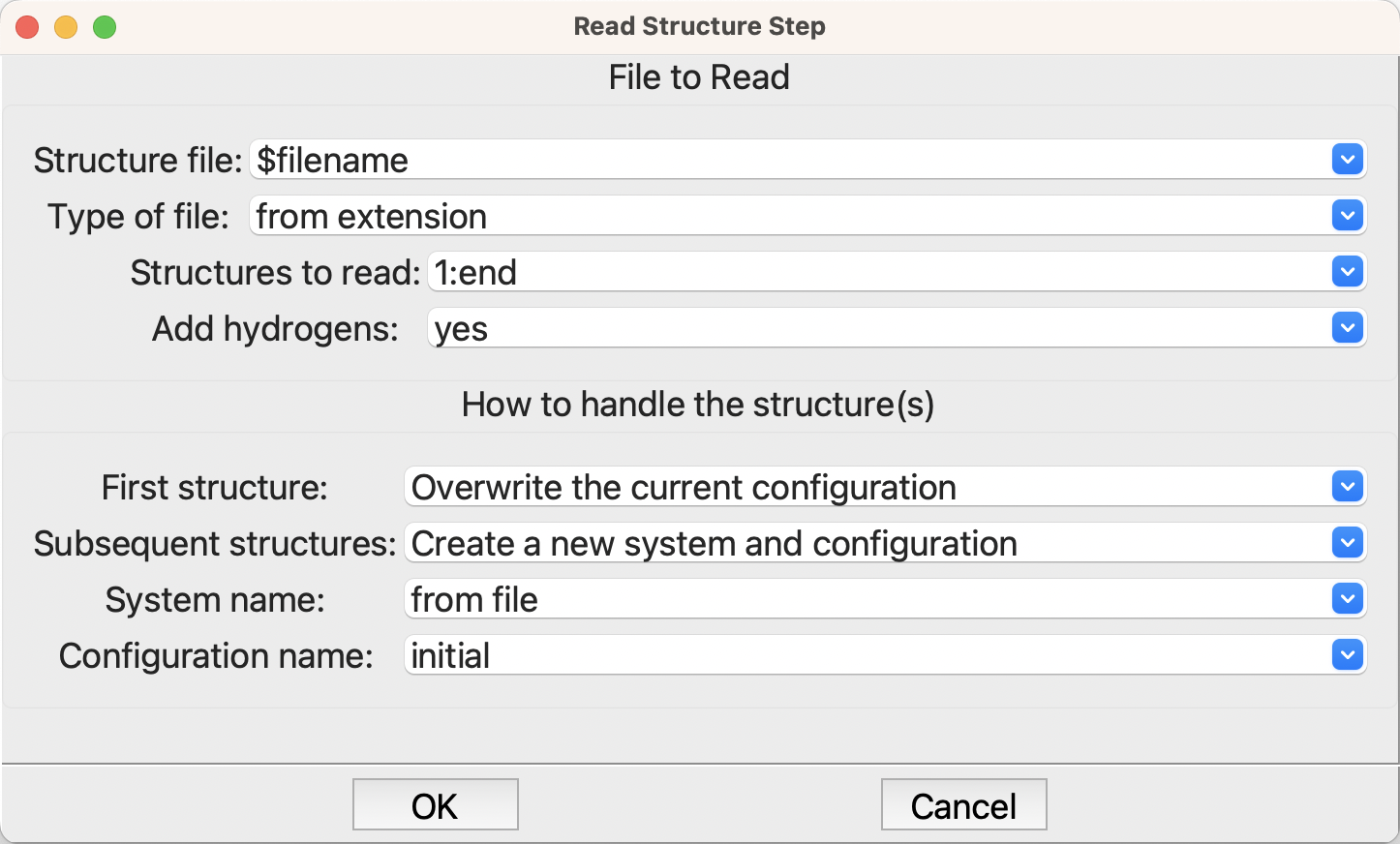
Creating and editing the parameters#
You can simply type the filename into the first entry; however, it is usually better to use the Control Parameters step to read a filename when the job is run, as in this example. This approach lets you easily run different molecules.
Normally you should let the step work out the type of file from the extension, rather than fix it to a particular type of file. However, if the code doesn’t understand the type of file you should specify the type here.
This plug-in uses OpenBabel to read most types of files, but specializes the handling for some file types to improve on OpenBabel’s capabilities. This step can read most formats, so it is recommended to use the correct file extensions and let the code do the rest.
Some file formats can store multipl structures, and some do not include hydrogen atoms, so the next two options cover those cases. They have good defaults so you can usually ignore them.
Finally, you need to indicate where to put the structures and how to name them in SEAMM. Again the defaults are a reasonable place to start.
That should be enough to get started. For more detail about the functionality in this plug-in, see the User Guide.