Getting Started#
Installation#
The Forcefield step is probably already installed in your SEAMM environment, but if not or if you wish to check, follow the directions for the SEAMM Installer. The graphical installer is the easiest to use. In the SEAMM conda environment, simply type:
seamm-installer
or use the shortcut if you installed one. Switch to the second tab, Components, and check for forcefield-step. If it is not installed, or can be updated, check the box next to it and click Install selected or Update selected as appropriate.
The non-graphical installer is also straightforward:
seamm-installer install --update forcefield-step
will ensure both that it is installed and up-to-date.
Intoduction to the Forcefield Step#
The Forcefield Step is used to read a forcefield into SEAMM and set it as the default forcefield until another Forcefield Step changes the default. You can also use the Forcefield Step to assign the forcefield to your system, though the assignment will also be done automatically when needed so you do not need to explicitly ask the Forcefield Step to do the assignment.
The Forcefield Step resides in the Simulation section of the Flowchart. Typically it is one of the firt couple steps in a Flowchart, defining the forcefield to be used by subsequent steps:
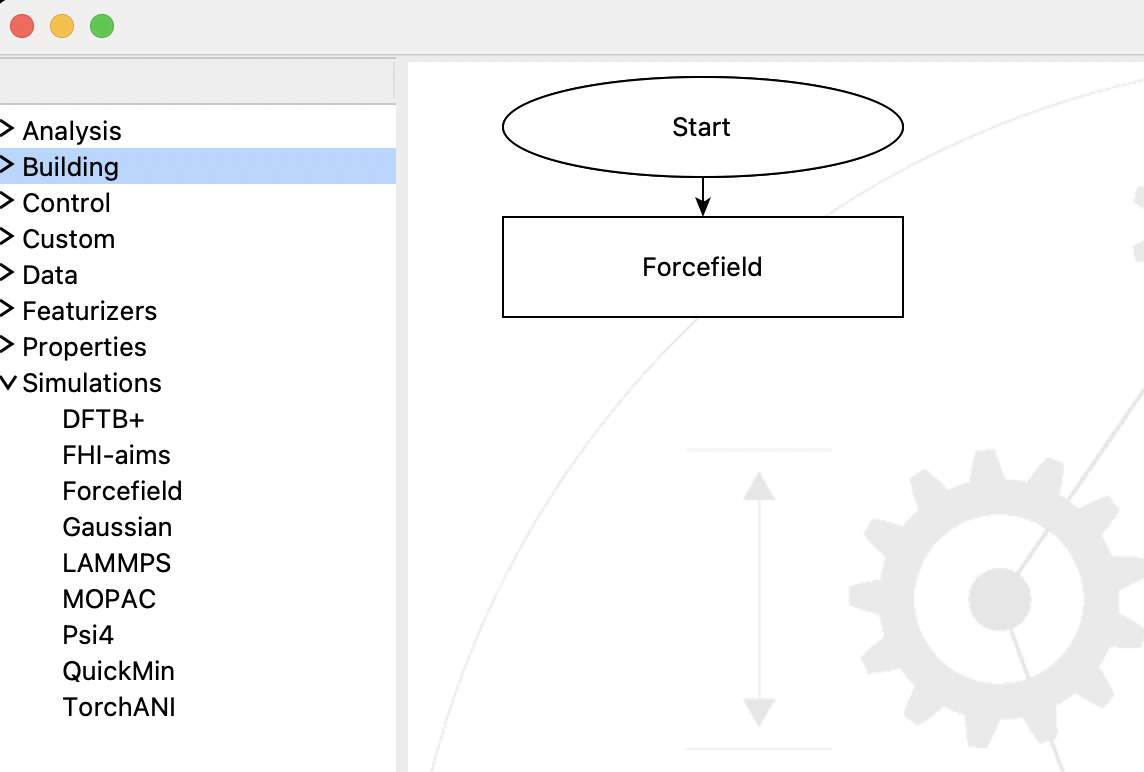
A flowchart with a single Forcefield Step#
Opening the Forcefield Step gives this dialog:
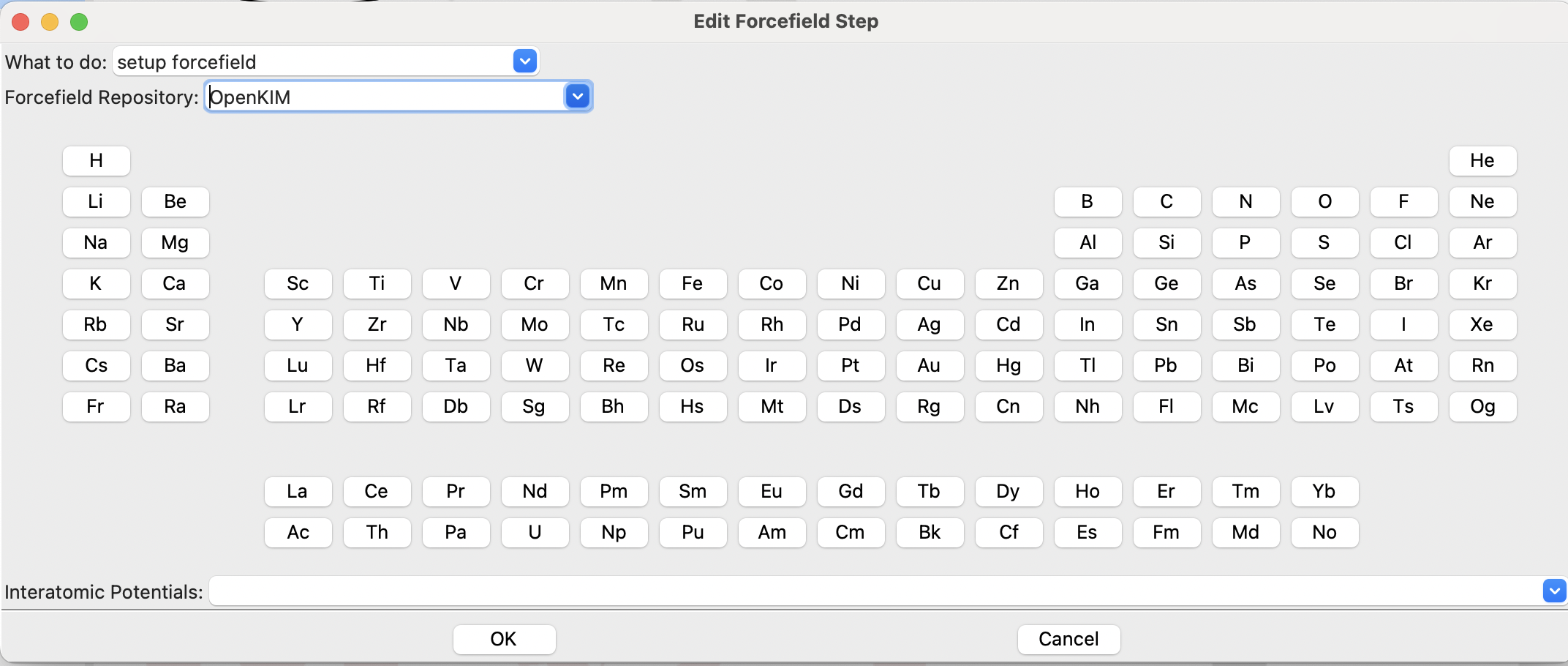
The initial forcefield dialog#
The first field, “What to do:”, allows you to setup a forcefield or to assign it to the current system, as mentioned above. The second, “Forcefield Repository”, is used to select the type of forcefield to setup:
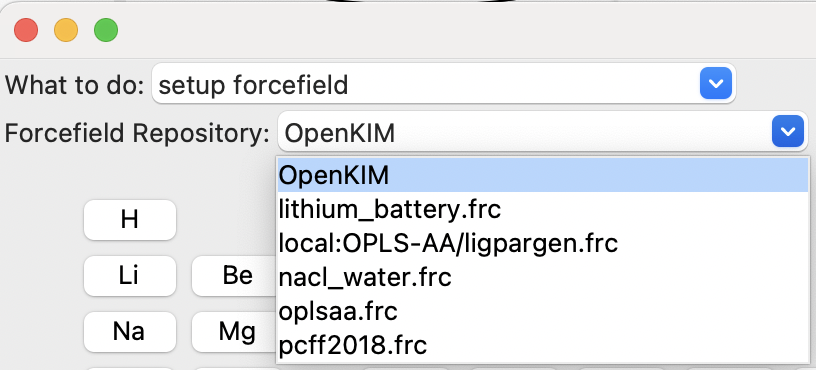
The choice of forcefield to set up.#
There are currently four main possibilities:
OpenKIM is an online repository that contains many of the “interatomic potentials” used for materials simulations. These are potentials such as the Stillinger-Webber potential for silicon, the potentials for the embedded atom method (EAM), and those for the modified embedded atom method (MEAM).
OPLS-AA, the Optimized Potentials for Liquid Simulations – All Atoms forcefield of William Jorgensen and et al. [1], has wide coverage of organic and biomolecular systems.
PCFF2018, the Polymer Consistent Force Field [2],also covers organic molecules with an emphasis on (synthetic) polymers.
local:OPLS_AA/ligpargen.frc contains parameters that the user has downloaded from the LigParGen server. See the The LigParGen Server section in the User Guide for more information.
Your installation may have a different set of forcefields than shown above. Once you have setup a forcefield you can proceed to e.g. LAMMPS to run the simulation using the forcefield.
That should be enough to get started. For more detail about the functionality in this plug-in, see the User Guide.